Naive Bayes¶
Class Reference¶
-
class
pykitml.NaiveBayes(input_size, output_size, distributions, reg_param=1)¶ Implements Naive Bayes classifier.
Note
Consider using
GaussianNaiveBayesif all of your features are continuous.-
__init__(input_size, output_size, distributions, reg_param=1)¶ Parameters: - input_size (int) – Size of input data or number of input features.
- output_size (int) – Number of categories or groups.
- distribution (list) – List of strings describing the distribution to use
for each feature. Option are
'gaussian','binomial','multinomial'. - reg_param (int) – If a given class and feature value never occur together in the training data,
then the frequency-based probability estimate will be zero.
This is problematic because it will wipe out all information in the other
probabilities when they are multiplied.
So, the probability will become
log(reg_param). This is a way to regularize Naive Bayes classifier. See https://en.wikipedia.org/wiki/Naive_Bayes_classifier#Multinomial_naive_Bayes
Raises: InvalidDistributionType– If invalid distribution. Can only be'gaussian','binomial','multinomial'.IndexError– If the input_size does not match the length of distribution length.
-
feed(input_data)¶ Accepts input array and feeds it to the model.
Parameters: input_data (numpy.array) – The input to feed the model. Raises: ValueError– If the input data has invalid dimensions/shape.Note
This function only feeds the input data, to get the output after calling this function use
get_output()orget_output_onehot()
-
get_output()¶ Returns the output activations of the model.
Returns: The output activations. Return type: numpy.array
-
get_output_onehot()¶ Returns the output layer activations of the model as a one-hot array. A one-hot array is an array of bits in which only one of the bits is high/true. In this case, the corresponding bit to the neuron/node having the highest activation will be high/true.
Returns: The one-hot output activations array. Return type: numpy.array
-
train(training_data, targets)¶ Trains the model on the training data.
Parameters: - training_data (numpy.array) – numpy array containing training data.
- targets (numpy.array) – numpy array containing training targets, corresponding to the training data.
Raises: numpy.AxisError– If output_size is less than two. Usepykitml.onehot()to change 0/False to [1, 0] and 1/True to [0, 1] for binary classification.
-
accuracy(testing_data, testing_targets)¶ Tests the accuracy of the model on the testing data passed to the function. This function should be only used for classification.
Parameters: - testing_data (numpy.array) – numpy array containing testing data.
- testing_targets (numpy.array) – numpy array containing testing targets, corresponding to the testing data.
Returns: accuracy – The accuracy of the model over the testing data i.e how many testing examples did the model predict correctly.
Return type: float
-
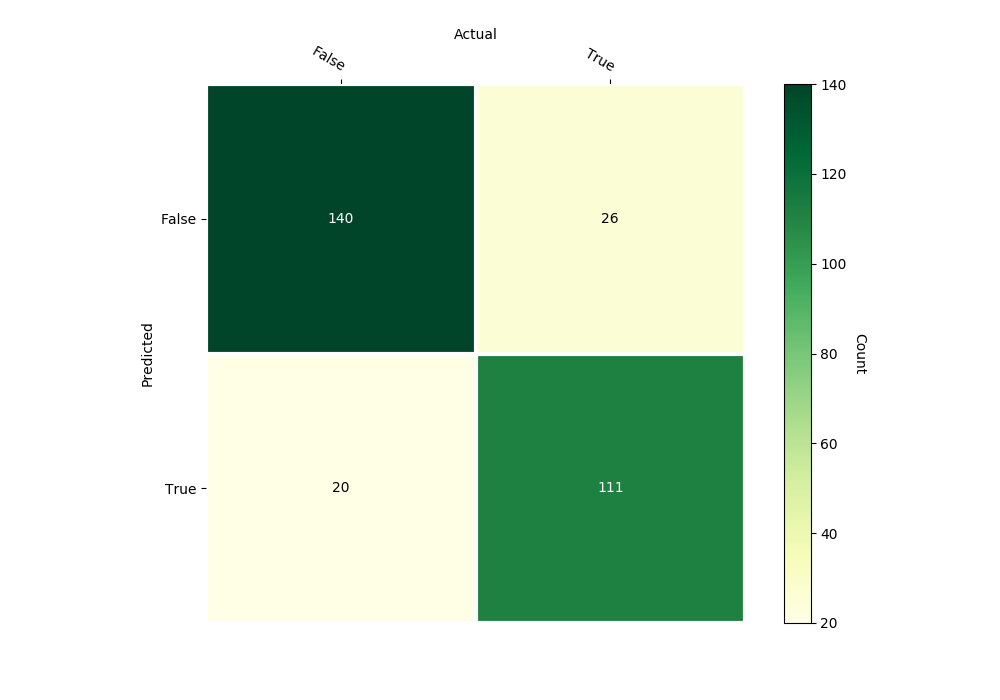
confusion_matrix(test_data, test_targets, gnames=[], plot=True)¶ Returns and plots confusion matrix on the given test data.
Parameters: - test_data (numpy.array) – Numpy array containing test data
- test_targets (numpy.array) – Numpy array containing the targets corresponding to the test data.
- plot (bool) – If set to false, will not plot the matrix. Default is true.
- gnames (list) – List of string names for each class/group.
Returns: confusion_matrix – The confusion matrix.
Return type: numpy.array
-
Example: Heart Disease Prediction¶
Dataset
Heart Disease - pykitml.datasets.heartdisease module
Training
import os.path
import pykitml as pk
from pykitml.datasets import heartdisease
# Download the dataset
if not os.path.exists('heartdisease.pkl'):
heartdisease.get()
# Load heart data set
inputs, outputs = heartdisease.load()
# Change 0/False to [1, 0]
# Change 1/True to [0, 1]
outputs = pk.onehot(outputs)
distrbutions = [
'gaussian', 'binomial', 'multinomial',
'gaussian', 'gaussian', 'binomial', 'multinomial',
'gaussian', 'binomial', 'gaussian', 'multinomial',
'multinomial', 'multinomial'
]
# Create model
bayes_heart_classifier = pk.NaiveBayes(13, 2, distrbutions)
# Train
bayes_heart_classifier.train(inputs, outputs)
# Save it
pk.save(bayes_heart_classifier, 'bayes_heart_classifier.pkl')
# Print accuracy
accuracy = bayes_heart_classifier.accuracy(inputs, outputs)
print('Accuracy:', accuracy)
# Plot confusion matrix
bayes_heart_classifier.confusion_matrix(inputs, outputs,
gnames=['False', 'True'])
Predict heartdisease for a person with age, sex, cp, trestbps, chol, fbs, restecg, thalach, exang, oldpeak, slope, ca, thal: 67, 1, 4, 160, 286, 0, 2, 108, 1, 1.5, 2, 3, 3
import numpy as np
import pykitml as pk
# Predict heartdisease for a person with
# age sex cp trestbps chol fbs restecg thalach exang oldpeak slope ca thal
# 67, 1, 4, 160, 286, 0, 2, 108, 1, 1.5, 2, 3, 3
input_data = np.array([67, 1, 4, 160, 286, 0, 2, 108, 1, 1.5, 2, 3, 3], dtype=float)
# Load the model
bayes_heart_classifier = pk.load('bayes_heart_classifier.pkl')
# Get output
bayes_heart_classifier.feed(input_data)
model_output = bayes_heart_classifier.get_output()
# Print result (log of probabilities)
print(model_output)
Confusion Matrix